Family known  |
|||||||
| Total |
100%  |
<100%  |
Family unknown  |
||||
| Functional domains | 8239 | 5779 | 0 | 2460 | |||
| UniProtKB | 6 | 0 | 0 | 6 | |||
| GI | 14 | 0 | 0 | 14 | |||
| Structures | 25 | ||||||
| Reactions | 0 | ||||||
| Functional domains of this subgroup were last updated on June 25, 2017 | |||||||
| New functional domains were last added to this subgroup on Oct. 7, 2014 | |||||||
The Biotin and thiamin synthesis-associated domain (BATS, IPR010722) is found as part of the functional domain in all members of this subgroup. It is especially associated with Biotin Synthase (BioB) and 2-iminoacetate synthase (ThiH), the latter of which is associated with thiamin biosynthesis. It is not completely clear exactly what the role of this domain is yet, it maybe involved in co-factor binding or dimerisation. All members of this subgroup also have secondary iron-sulfur clusters.
Vey JL, Drennan CL.
Structural insights into radical generation by the radical SAM superfamily
▸ Abstract
Chem Rev. 2011;111(4):2487-2506 | PubMed ID: 21370834
Phalip V, Lemoine Y, Jeltsch JM.
Cloning of Schizosaccharomyces pombe bio2 by heterologous complementation of a Saccharomyces cerevisiae mutant
▸ Abstract
Curr Microbiol. 1999;39(6):348-350 | PubMed ID: 10525840
Lotierzo M, Tse Sum Bui B, Florentin D, Escalettes F, Marquet A.
Biotin synthase mechanism: an overview
▸ Abstract
Biochem Soc Trans. 2005;33(4):820-823 | PubMed ID: 16042606
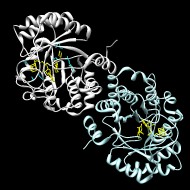
Quitterer F, List A, Eisenreich W, Bacher A, Groll M.
Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis
▸ Abstract
Angew Chem Int Ed Engl. 2012;51(6):1339-1342 | PubMed ID: 22095926
Nicolet Y, Rubach JK, Posewitz MC, Amara P, Mathevon C, Atta M, Fontecave M, Fontecilla-Camps JC.
X-ray structure of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima
▸ Abstract
J Biol Chem 2008;283(27):18861-18872 | PubMed ID: 18400755
Family members of this subgroup include:
Biotin synthase (BioB) catalyses the final step in the biosynthesis of the cofactor biotin. It requires both S-adenosylmethionine, a [4Fe-4S] and a [2Fe-2S] cluster. The [2Fe-2S] cluster is the source of the sulfur that is inserted between the C6 and C8 of dithiobiotin. BioB utilises two molecules of SAM for each catalytic turnover and is stoichiometric in its use of SAM. AN electron transfer system is also required for catalytic activity. In E. coli, this is NADPH, flavodoxin and flavodoxin reductase.
HmdB represents a family of enzymes that are involved in the construction of the metallo-cofactor HmdA. They do not have the canonical CxxxCxxC motif, instead having a CX5CX2C motif.
HydE represents a family of enzymes that are involved in the maturation of the [FeFe]-hydrogenase active site, the exact role of this family of proteins is unknown, bur thought to be involved in the formation and incorporation of the -C#N and C#O ligands.
PylB (methylornithine synthase) represents a family of enzymes that are responsible for the first enzyme in the synthesis of pyrrolysine.
ThiH
NosL
HydG
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| repnet.sg1060.th50.pE20.mek250.xgmml | Representative network: each node is a group of similar sequences | node similarity threshold = 50 max edge count = 250 min -log10 E = 20 |
size = 9.2M num_edges = 13744 num_nodes = 285 |
| sfld_alignment_sg1060.msa | Annotated Sequence Alignment, Stockholm format | 254 sequences size: 159K |
| Subgroup ▸ Legend | T  |
K  |
C  |
U  |
S  |
||
|---|---|---|---|---|---|---|---|
| BATS domain containing | 8239 | 5779 | 5779 | 2460 | 25 | ||
| ┗ 1: biotin synthase like | 5006 | 3245 | 3245 | 1761 | 1 | ||
| ┗ biotin synthase | 3245 | 3245 | 3245 | 1 | |||
| ┗ 1: cleavage of the Ca-Cb bond in aromatic amino acids | 2304 | 2231 | 2231 | 73 | 4 | ||
| ┗ 2-iminoacetate synthase (ThiH) | 1523 | 1523 | 1523 | 0 | |||
| ┗ 3-methyl-2-indolic acid synthase (NosL) | 5 | 5 | 5 | 0 | |||
| ┗ [Fe] hydrogenase maturase (HydG-like) | 703 | 703 | 703 | 0 | |||
| ┗ 1: HMD cofactor maturase (HmdB-like) | 39 | 39 | 39 | 0 | 0 | ||
| ┗ HMD cofactor maturase (HmdB-like) | 39 | 39 | 39 | 0 | |||
| ┗ 1: HydE/PylB-like | 886 | 264 | 264 | 622 | 20 | ||
| ┗ 3-methylornithine synthase (PylB-like) | 42 | 42 | 42 | 1 | |||
| ┗ [FeFe] hydrogenase maturase (HydE-like) | 222 | 222 | 222 | 10 |