| Total |
100%  |
<100%  |
|||
| Functional domains | 42 | 42 | 0 | ||
| UniProtKB | 48 | 48 | 0 | ||
| GI | 179 | 179 | 0 | ||
| Structures | 1 | ||||
| Reactions | 1 | ||||
| Functional domains of this family were last updated on June 10, 2017 | |||||
| New functional domains were last added to this family on June 22, 2014 | |||||
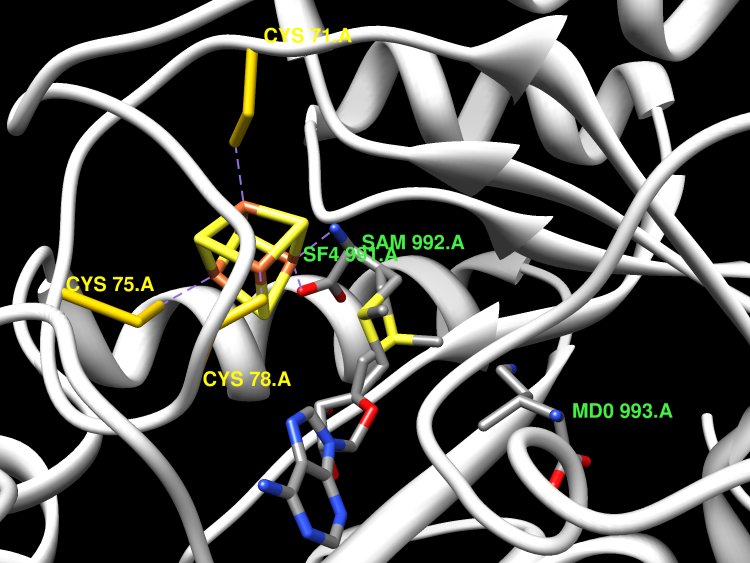
Pyrrolysine is the 22nd naturally occurring amino acid residue and is formed from two lysine molecules in a reaction pathway that requires three proteins: PylB, PylC and PylD. PylB, the first enzyme in the pathway, shares the CxxxCxxC canonical [4Fe-4S] binding motif that marks it as a member of the radical SAM superfamily. PylB is responsible for the isomerisation of L-lysine into (2R,3R) 3-methylornithine. It is currently unclear if PylB is a single-turnover enzyme, studies have shown that, at least in the absence of PylC and/or PylD, it is only capable of a single-turnover.
Quitterer F, List A, Eisenreich W, Bacher A, Groll M.
Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis
▸ Abstract
Angew Chem Int Ed Engl. 2012;51(6):1339-1342 | PubMed ID: 22095926
See http://www.sciencedirect.com/science/article/pii/S1367593113001233 Fig 4 for proposed mechanism review. and http://onlinelibrary.wiley.com/doi/10.1002/anie.201106765/full Scheme 2 for original proposal
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| network.fam349.bs60.mek250K.xgmml | One node per sequence network | min bit score = 60 max edge count = 250K |
size = 508K num_edges = 861 num_nodes = 42 |
| sfld_alignment_fam349.msa | Annotated Sequence Alignment, Stockholm format | 14 sequences size: 7.0K |
Active Site