| Total |
100%  |
<100%  |
|||
| Functional domains | 660 | 659 | 1 | ||
| UniProtKB | 633 | 632 | 1 | ||
| GI | 1430 | 1427 | 3 | ||
| Structures | 0 | ||||
| Reactions | 6 | ||||
| Functional domains of this family were last updated on June 10, 2017 | |||||
| New functional domains were last added to this family on June 22, 2014 | |||||
Involved in the C2 methylation of bacterial hopanoids. Although the reaction is shown with methylcobalamin acting as the methyl donor, it is possible that SAM might be the methyl donor for the reaction (as this is another function of the SAM cofactor).
Welander PV, Coleman ML, Sessions AL, Summons RE, Newman DK
Identification of a methylase required for 2-methylhopanoid production and implications for the interpretation of sedimentary hopanes
▸ Abstract
Proc Natl Acad Sci U S A 2010;107(19):8537-8542 | PubMed ID: 20421508
No notes.
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| network.fam303.bs60.mek250K.xgmml | One node per sequence network | min bit score = 60 max edge count = 250K |
size = 110M num_edges = 212747 num_nodes = 660 |
| sfld_alignment_fam303.msa | Annotated Sequence Alignment, Stockholm format | 169 sequences size: 103K |
Catalyzed Reaction(s)
hop-22(29)-ene C2-methyltransferase
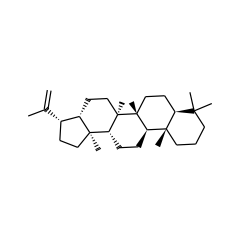 |
+ 2 |  |
 |
 |
+ | 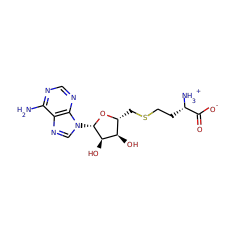 |
+ |  |
+ |  |
||
| hop-22(29)-ene 4648 |
S-adenosyl-L-methionine zwitterion 59789 |
2-methylhop-22(29)-ene 132460 |
S-adenosyl-L-homocysteine zwitterion 57856 |
L-methionine zwitterion 57844 |
5'-deoxyadenosine 17319 |
EC: | IntEnz: | Kegg: | BioCyc: | BRENDA: |
hopan-22-ol 2-methyltransferase
 |
+ 2 |  |
 |
 |
+ |  |
+ |  |
+ |  |
||
| hopan-22-ol 36484 |
S-adenosyl-L-methionine zwitterion 59789 |
2-methyl-hopan-22-ol 132462 |
S-adenosyl-L-homocysteine zwitterion 57856 |
L-methionine zwitterion 57844 |
5'-deoxyadenosine 17319 |
EC: | IntEnz: | Kegg: | BioCyc: | BRENDA: |
tetrahymanol 2-methyltransferase
 |
+ 2 |  |
 |
 |
+ |  |
+ |  |
+ |  |
||
| tetrahymanol 9493 |
S-adenosyl-L-methionine zwitterion 59789 |
2-methyltetrahymanol 132463 |
S-adenosyl-L-homocysteine zwitterion 57856 |
L-methionine zwitterion 57844 |
5'-deoxyadenosine 17319 |
EC: | IntEnz: | Kegg: | BioCyc: | BRENDA: |
hop-21-ene 2-methyltransferase
 |
+ 2 |  |
 |
 |
+ |  |
+ |  |
+ |  |
||
| hop-21-ene 132464 |
S-adenosyl-L-methionine zwitterion 59789 |
2-methylhop-21-ene 132465 |
S-adenosyl-L-homocysteine zwitterion 57856 |
L-methionine zwitterion 57844 |
5'-deoxyadenosine 17319 |
EC: | IntEnz: | Kegg: | BioCyc: | BRENDA: |
hop-17(21)-ene 2-methyltransferase
 |
+ 2 |  |
 |
 |
+ |  |
+ |  |
+ |  |
||
| hop-17(21)-ene 132466 |
S-adenosyl-L-methionine zwitterion 59789 |
2-methylhop-17(21)-ene 132467 |
S-adenosyl-L-homocysteine zwitterion 57856 |
L-methionine zwitterion 57844 |
5'-deoxyadenosine 17319 |
EC: | IntEnz: | Kegg: | BioCyc: | BRENDA: |
bacteriohopanetetrol 2-methyltransferase
 |
+ 2 |  |
 |
 |
+ |  |
+ |  |
+ |  |
||
| bacteriohopanetetrol 132468 |
S-adenosyl-L-methionine zwitterion 59789 |
2-methylbacteriohopanetetrol 132469 |
S-adenosyl-L-homocysteine zwitterion 57856 |
L-methionine zwitterion 57844 |
5'-deoxyadenosine 17319 |
EC: | IntEnz: | Kegg: | BioCyc: | BRENDA: |


