Top Level Name
⌊ Superfamily (core) Haloacid Dehalogenase
⌊ Subgroup C1.8: polynucleotide 5'-hydroxyl-kinase c-terminal phosphatase like
Family known  |
|||||||
| Total |
100%  |
<100%  |
Family unknown  |
||||
| Functional domains | 58 | 0 | 1 | 57 | |||
| UniProtKB | 86 | 0 | 1 | 85 | |||
| GI | 209 | 0 | 4 | 205 | |||
| Structures | 5 | ||||||
| Reactions | 0 | ||||||
| Functional domains of this subgroup were last updated on Nov. 22, 2017 | |||||||
| New functional domains were last added to this subgroup on Aug. 1, 2014 | |||||||
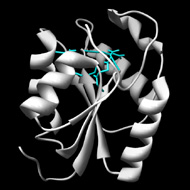
Structurally characterized enzymes in this subgroup have the C1 cap type. Functionally characterized enzymes are polynucleotide 5'-hydroxyl-kinase c-terminal phosphatases. A divalent metal ion cofactor is likely required for activity.
No References.
No notes.
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| network.sg10.bs60.mek250K.xgmml | One node per sequence network | min bit score = 60 max edge count = 250K |
size = 765K num_edges = 1327 num_nodes = 58 |
| repnet.sg10.th50.pE20.mek250.xgmml | Representative network: each node is a group of similar sequences | node similarity threshold = 50 max edge count = 250 min -log10 E = 20 |
size = 95K num_edges = 128 num_nodes = 22 |
| sfld_alignment_sg10.msa | Annotated Sequence Alignment, Stockholm format | 2 sequences size: 1.4K |
| Subgroup ▸ Legend | T  |
K  |
C  |
U  |
S  |
||
|---|---|---|---|---|---|---|---|
| C1.8: polynucleotide 5'-hydroxyl-kinase c-terminal phosphatase like | 58 | 1 | 0 | 57 | 5 | ||
| ┗ polynucleotide 5'-hydroxyl-kinase c-terminal phosphatase | 1 | 1 | 0 | 5 |