Hänzelmann P, Schindelin H
Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
▸ Abstract
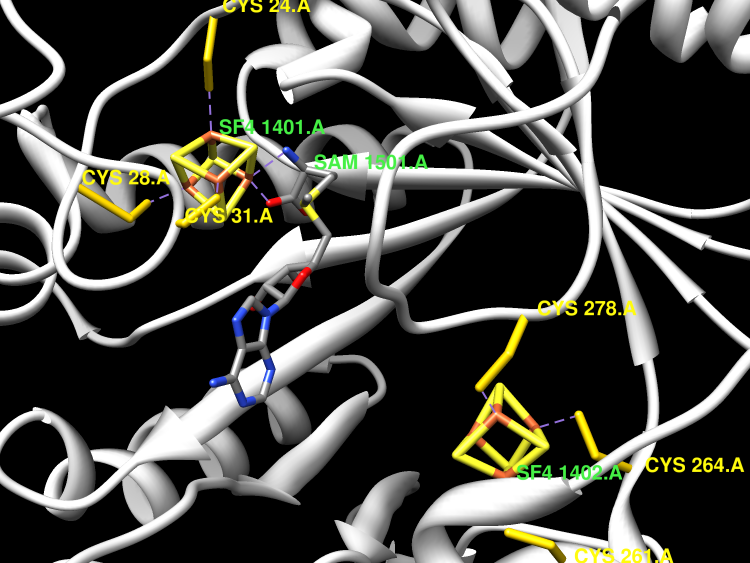
The first step in molybdenum cofactor biosynthesis, the conversion of 5'-GTP to precursor Z, an oxygen-sensitive tetrahydropyranopterin is catalyzed by the S-adenosylmethionine (SAM)-dependent enzyme MoaA and the accessory protein MoaC. This reaction involves the radical-initiated intramolecular rearrangement of the guanine C8 atom. MoaA harbors an N-terminal [4Fe-4S] cluster, which is involved in the reductive cleavage of SAM and generates a 5'-deoxyadenosyl radical (5'-dA*), and a C-terminal [4Fe-4S] cluster presumably involved in substrate binding and/or activation. Biochemical studies identified residues involved in 5'-GTP binding and the determinants of nucleotide specificity. The crystal structure of MoaA in complex with 5'-GTP confirms the biochemical data and provides valuable insights into the subsequent radical reaction. MoaA binds 5'-GTP with high affinity and interacts through its C-terminal [4Fe-4S] cluster with the guanine N1 and N2 atoms, in a yet uncharacterized binding mode. The tightly anchored triphosphate moiety prevents the escape of radical intermediates. This structure also visualizes the L-Met and 5'-dA cleavage products of SAM. Rotation of the 5'-dA ribose and/or conformational changes of the guanosine are proposed to bring the 5'-deoxyadenosyl radical into close proximity of either the ribose C2' and C3' or the guanine C8 carbon atoms leading to hydrogen abstraction.
Proc Natl Acad Sci U S A
2006;103(18):6829-6834
| PubMed ID:
16632608
Hänzelmann P, Schindelin H
Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans
▸ Abstract
The MoaA and MoaC proteins catalyze the first step during molybdenum cofactor biosynthesis, the conversion of a guanosine derivative to precursor Z. MoaA belongs to the S-adenosylmethionine (SAM)-dependent radical enzyme superfamily, members of which catalyze the formation of protein and/or substrate radicals by reductive cleavage of SAM by a [4Fe-4S] cluster. A defined in vitro system is described, which generates precursor Z and led to the identification of 5'-GTP as the substrate. The structures of MoaA in the apo-state (2.8 angstroms) and in complex with SAM (2.2 angstroms) provide valuable insights into its mechanism and help to define the defects caused by mutations in the human ortholog of MoaA that lead to molybdenum cofactor deficiency, a usually fatal disease accompanied by severe neurological symptoms. The central core of each subunit of the MoaA dimer is an incomplete triosephosphate isomerase barrel formed by the N-terminal part of the protein, which contains the [4Fe-4S] cluster typical for SAM-dependent radical enzymes. SAM is the fourth ligand to the cluster and binds to its unique Fe as an N/O chelate. The lateral opening of the incomplete triosephosphate isomerase barrel is covered by the C-terminal part of the protein containing an additional [4Fe-4S] cluster, which is unique to MoaA proteins. Both FeS clusters are separated by approximately 17 angstroms, with a large active site pocket between. The noncysteinyl-ligated unique Fe site of the C-terminal [4Fe-4S] cluster is proposed to be involved in the binding and activation of 5'-GTP.
Proc Natl Acad Sci U S A
2004;101(35):12870-12875
| PubMed ID:
15317939
Hänzelmann P, Schwarz G, Mendel RR
Functionality of alternative splice forms of the first enzymes involved in human molybdenum cofactor biosynthesis
▸ Abstract
In humans, genetic deficiencies of enzymes involved in molybdenum cofactor biosynthesis trigger an autosomal recessive and usually fatal disease with severe mostly neurological symptoms. In each of the three biosynthesis steps, at least two proteins or domains are linked for catalysis. For steps 1 and 2, bicistronic mocs (molybdenum cofactor synthesis) mRNAs were found (mocs1 and mocs2) that have been proposed to encode two separate proteins (A and B). In both cases, the A proteins share a highly conserved ubiquitin-like double glycine motif, which is functionally important at least for the small subunit of molybdopterin (MPT) synthase (MOCS2A). Besides the bicistronic form of mocs1, two alternative splice transcripts were found, resulting in the expression of multidomain proteins embodying both MOCS1A, but without the double glycine motif, and the entire MOCS1B. Here we describe the first functional characterization of the human proteins MOCS1A and MOCS1B as well as the MOCS1A-MOCS1B fusion proteins that catalyze the formation of precursor Z, a 6-alkyl pterin with a cyclic phosphate, the immediate precursor of MPT in molybdenum cofactor biosynthesis. High level expression of MOCS1A and MOCS1B in Escherichia coli resulted in the formation and accumulation of precursor Z that was subsequently converted to MPT. We showed that for catalytic activity MOCS1A needs an accessible C-terminal double glycine motif. In the MOCS1A-MOCS1B fusion proteins lacking the MOCS1A double glycines, only MOCS1B activity could be detected. No evidence was found for an expression of MOCS1B from the bicistronic mocs1A-mocs1B splice type I cDNA, indicating that MOCS1B is only expressed as a fusion to an inactive MOCS1A. Comparative mutational studies of MOCS1A and the small subunit of the E. coli MPT synthase (MoaD) indicate a different function of the double glycine motifs in both proteins.
J Biol Chem.
2002;277(21):18303-18312
| PubMed ID:
11891227
Hänzelmann P, Hernández HL, Menzel C, García-Serres R, Huynh BH, Johnson MK, Mendel RR, Schindelin H
Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis
▸ Abstract
The human proteins MOCS1A and MOCS1B catalyze the conversion of a guanosine derivative to precursor Z during molybdenum cofactor biosynthesis. MOCS1A shares homology with S-adenosylmethionine (AdoMet)-dependent radical enzymes, which catalyze the formation of protein and/or substrate radicals by reductive cleavage of AdoMet through a [4Fe-4S] cluster. Sequence analysis of MOCS1A showed two highly conserved cysteine motifs, one near the N terminus and one near the C terminus. MOCS1A was heterologously expressed in Escherichia coli and purified under aerobic and anaerobic conditions. Individual mutations of the conserved cysteines to serine revealed that all are essential for synthesis of precursor Z in vivo. The type and properties of the iron-sulfur (FeS) clusters were investigated using a combination of UV-visible absorption, variable temperature magnetic circular dichroism, resonance Raman, Mössbauer, and EPR spectroscopies coupled with iron and acid-labile sulfide analyses. The results indicated that anaerobically purified MOCS1A is a monomeric protein containing two oxygen-sensitive FeS clusters, each coordinated by only three cysteine residues. A redox-active [4Fe-4S](2+,+) cluster is ligated by an N-terminal CX(3)CX(2)C motif as is the case with all other AdoMet-dependent radical enzymes investigated thus far. A C-terminal CX(2)CX(13)C motif that is unique to MOCS1A and its orthologs primarily ligates a [3Fe-4S](0) cluster. However, MOCS1A could be reconstituted in vitro under anaerobic conditions to yield a form containing two [4Fe-4S](2+) clusters. The N-terminal [4Fe-4S](2+) cluster was rapidly degraded by oxygen via a semistable [2Fe-2S](2+) cluster intermediate, and the C-terminal [4Fe-4S](2+) cluster was rapidly degraded by oxygen to yield a semistable [3Fe-4S](0) cluster intermediate.
J Biol Chem.
2004;279(33):34721-34732
| PubMed ID:
15180982
Mehta AP, Hanes JW, Abdelwahed SH, Hilmey DG, Hänzelmann P, Begley TP
Catalysis of a new ribose carbon-insertion reaction by the molybdenum cofactor biosynthetic enzyme MoaA
▸ Abstract
MoaA, a radical S-adenosylmethionine enzyme, catalyzes the first step in molybdopterin biosynthesis. This reaction involves a complex rearrangement in which C8 of guanosine triphosphate is inserted between C2' and C3' of the ribose. This study identifies the site of initial hydrogen atom abstraction by the adenosyl radical and advances a mechanistic proposal for this unprecedented reaction.
Biochemistry
2013;52(7):1134-1136
| PubMed ID:
23286307
Hover BM, Loksztejn A, Ribeiro AA, Yokoyama K
Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis
▸ Abstract
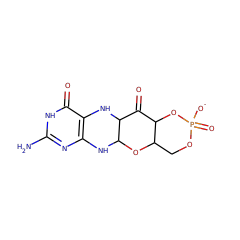
The molybdenum cofactor (Moco) is a redox cofactor found in all kingdoms of life, and its biosynthesis is essential for survival of many organisms, including humans. The first step of Moco biosynthesis is a unique transformation of guanosine 5'-triphosphate (GTP) into cyclic pyranopterin monophosphate (cPMP). In bacteria, MoaA and MoaC catalyze this transformation, although the specific functions of these enzymes were not fully understood. Here, we report the first isolation and structural characterization of a product of MoaA. This molecule was isolated under anaerobic conditions from a solution of MoaA incubated with GTP, S-adenosyl-L-methionine, and sodium dithionite in the absence of MoaC. Structural characterization by chemical derivatization, MS, and NMR spectroscopy suggested the structure of this molecule to be (8S)-3',8-cyclo-7,8-dihydroguanosine 5'-triphosphate (3',8-cH2GTP). The isolated 3',8-cH2GTP was converted to cPMP by MoaC or its human homologue, MOCS1B, with high specificities (Km < 0.060 μM and 0.79 ± 0.24 μM for MoaC and MOCS1B, respectively), suggesting the physiological relevance of 3',8-cH2GTP. These observations, in combination with some mechanistic studies of MoaA, unambiguously demonstrate that MoaA catalyzes a unique radical C-C bond formation reaction and that, in contrast to previous proposals, MoaC plays a major role in the complex rearrangement to generate the pyranopterin ring.
J Am Chem Soc
2013;135(18):7019-7032
| PubMed ID:
23627491
Mehta AP, Abdelwahed SH, Xu H, Begley TP
Molybdopterin Biosynthesis: Trapping of intermediates for the MoaA-catalyzed reaction using 2'-deoxyGTP and 2'- chloroGTP as substrate analogs
▸ Abstract
MoaA is a radical S-adenosylmethionine (AdoMet) enzyme that catalyzes a complex rearrangement of GTP in the first step of molybdopterin biosynthesis. In this paper, we provide additional characterization of the MoaA reaction product, describe the use of 2'-chloroGTP to trap the GTP C3' radical, generated by hydrogen atom transfer to the AdoMet-derived adenosyl radical, and the use of 2'-deoxyGTP to block a late step in the reaction sequence. These probes, coupled with the previously reported trapping of an intermediate in which C3' of the ribose is linked to C8 of the purine4, allow us to propose a plausible mechanism for the MoaA-catalyzed reaction.
J Am Chem Soc
2014;None(None):None-None
| PubMed ID:
24955657
Hover BM, Yokoyama K
C-terminal glycine-gated radical initiation by GTP 3',8-cyclase in the molybdenum cofactor biosynthesis
▸ Abstract
The molybdenum cofactor (Moco) is an essential redox cofactor found in all kingdoms of life. Genetic mutations in the human Moco biosynthetic enzymes lead to a fatal metabolic disorder, Moco deficiency (MoCD). Greater than 50% of all human MoCD patients have mutations in MOCS1A, a radical S-adenosyl-l-methionine (SAM) enzyme involved in the conversion of guanosine 5'-triphosphate (GTP) into cyclic pyranopterin monophosphate. In MOCS1A, one of the frequently affected locations is the GG motif constituted of two consecutive Gly at the C-terminus. The GG motif is conserved among all MOCS1A homologues, but its role in catalysis or the mechanism by which its mutation causes MoCD was unknown. Here, we report the functional characterization of the GG motif using MoaA, a bacterial homologue of MOCS1A, as a model. Our study elucidated that the GG motif is essential for the activity of MoaA to produce 3',8-cH2GTP from GTP (GTP 3',8-cyclase), and that synthetic peptides corresponding to the C-terminal region of wt-MoaA rescue the GTP 3',8-cyclase activity of the GG-motif mutants. Further biochemical characterization suggested that the C-terminal tail containing the GG motif interacts with the SAM-binding pocket of MoaA, and is essential for the binding of SAM and subsequent radical initiation. In sum, these observations suggest that the C-terminal tail of MoaA provides an essential mechanism to trigger the free radical reaction, impairment of which results in the complete loss of catalytic function of the enzyme, and causes MoCD.
J Am Chem Soc
2015;137(9):3352-3359
| PubMed ID:
25697423
Hover BM, Tonthat NK, Schumacher MA, Yokoyama K
Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis
▸ Abstract
The molybdenum cofactor (Moco) is essential for all kingdoms of life, plays central roles in various biological processes, and must be biosynthesized de novo. During Moco biosynthesis, the characteristic pyranopterin ring is constructed by a complex rearrangement of guanosine 5'-triphosphate (GTP) into cyclic pyranopterin (cPMP) through the action of two enzymes, MoaA and MoaC (molybdenum cofactor biosynthesis protein A and C, respectively). Conventionally, MoaA was considered to catalyze the majority of this transformation, with MoaC playing little or no role in the pyranopterin formation. Recently, this view was challenged by the isolation of 3',8-cyclo-7,8-dihydro-guanosine 5'-triphosphate (3',8-cH2GTP) as the product of in vitro MoaA reactions. To elucidate the mechanism of formation of Moco pyranopterin backbone, we performed biochemical characterization of 3',8-cH2GTP and functional and X-ray crystallographic characterizations of MoaC. These studies revealed that 3',8-cH2GTP is the only product of MoaA that can be converted to cPMP by MoaC. Our structural studies captured the specific binding of 3',8-cH2GTP in the active site of MoaC. These observations provided strong evidence that the physiological function of MoaA is the conversion of GTP to 3',8-cH2GTP (GTP 3',8-cyclase), and that of MoaC is to catalyze the rearrangement of 3',8-cH2GTP into cPMP (cPMP synthase). Furthermore, our structure-guided studies suggest that MoaC catalysis involves the dynamic motions of enzyme active-site loops as a way to control the timing of interaction between the reaction intermediates and catalytically essential amino acid residues. Thus, these results reveal the previously unidentified mechanism behind Moco biosynthesis and provide mechanistic and structural insights into how enzymes catalyze complex rearrangement reactions.
Proc Natl Acad Sci U S A
2015;112(20):6347-6352
| PubMed ID:
25941396