Top Level Name
⌊ Superfamily (core) Enolase
⌊ Subgroup muconate cycloisomerase
⌊ Family N-succinylamino acid racemase 2
| Total |
100%  |
<100%  |
|||
| Functional domains | 237 | 0 | 237 | ||
| UniProtKB | 376 | 0 | 376 | ||
| GI | 839 | 0 | 839 | ||
| Structures | 3 | ||||
| Reactions | 1 | ||||
| Functional domains of this family were last updated on Nov. 22, 2017 | |||||
| New functional domains were last added to this family on May 7, 2015 | |||||
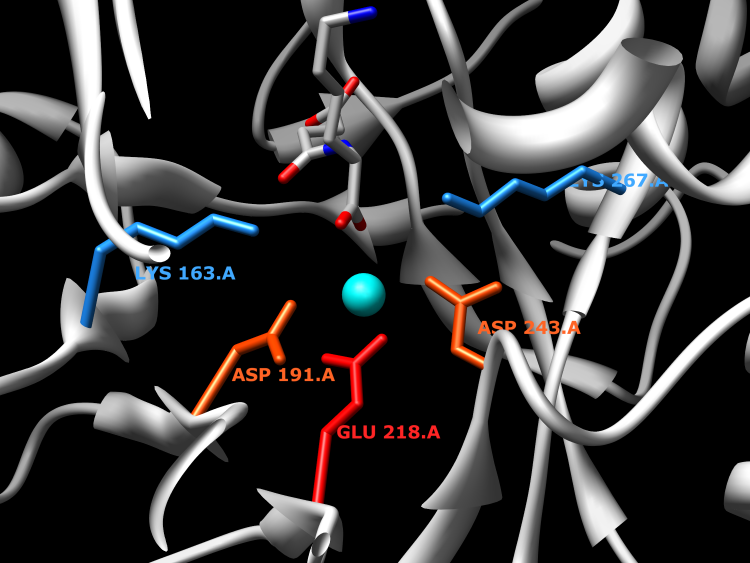
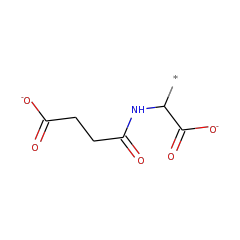
Enzymes in the N-succinylamino acid racemase 2 (NSAR2) family catalyze the racemization of N-succinyl arginine/lysine. Enzymes in this family are highly similar to enzymes in the dipeptide epimerase family. The experimentally characterized enzyme from Bacillus cereus ATCC 14579 has been shown to catalyze the epimerization of certain dipeptides, but at a substantially lower rate than the racemization of N-succinyl arginine/lysine. Although the reaction catalyzed by this family is similar to that catalyzed by the NSAR family (differing only in the preference of N-succinyl arginine/lysine for NSAR2 versus N-succinyl hydrophobic amino acids for NSAR), phylogenetic analysis suggests that these two families have independent evolutionary origins within the enolase superfamily. Not surprisingly, the amino acids responsible for substrate recognition appear to differ between the two families.
Song L, Kalyanaraman C, Fedorov AA, Fedorov EV, Glasner ME, Brown S, Imker HJ, Babbitt PC, Almo SC, Jacobson MP, Gerlt JA
Prediction and assignment of function for a divergent N-succinyl amino acid racemase
▸ Abstract
Nat Chem Biol 2007;3(8):486-491 | PubMed ID: 17603539
No notes.
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| network.fam154.bs60.mek250K.xgmml | One node per sequence network | min bit score = 60 max edge count = 250K |
size = 9.4M num_edges = 27966 num_nodes = 237 |
| sfld_alignment_fam154.msa | Annotated Sequence Alignment, Stockholm format | 3 sequences size: 2.7K |
Active Site