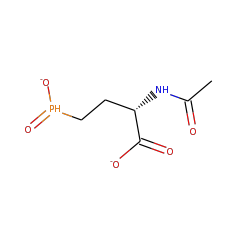
Top Level Name
⌊ Superfamily (core) Radical SAM
⌊ Subgroup B12-binding domain containing
⌊ Family P-methyltransferase (PhpK-like)
⌊ FunctionalDomain P-methyltransferase PhpK (ID 412962)
No Notes.
| Superfamily Assignment Evidence Code(s) | ISS |
| Family Assignment Evidence Code | CFM PubMed:21950770 |
| This entry was last updated on | June 10, 2017 |
References to Other Databases
Genbank
| Species | GI | Accession | Proteome |
|---|---|---|---|
| Streptomyces viridochromogenes Taxon ID: 1938 | 490086675 | WP_003988629.1 (RefSeq) | URP |
| Streptomyces viridochromogenes DSM 40736 Taxon ID: 591159 | 302467421 | EFL30514.1 (Genbank) | URP |
| Streptomyces viridochromogenes Taxon ID: 1938 | 51317960 | AAU00085.1 (Genbank) | URP |
| Streptomyces viridochromogenes Taxon ID: 1938 | 68697722 | URP | |
| obsolete GI = 302549803 | |||
| Show All | |||
Uniprot
| Protein Name | Accession | EC Number
 |
Identifier |
|---|---|---|---|
| n/a | Q5IW50 | Q5IW50_STRVR (TrEMBL) | |
| n/a | D9XF35 | D9XF35_STRVT (TrEMBL) |
Length of Enzyme (full-length): 549 | Length of Functional Domain: 540
MKHCIVVGYHETDMSGEELQLALEHGGDELPAPIRSMLRTKITFGGAHYSYLDALSEVRN
QDRGTDHAYTVSELPSLGTLYLVNYLQEQGHPADFVNSYTFEQDQLVKLLADNPASVAIT
TTFYMMPAPVIEIVRFIRRHNPSVPVIVGGPLVDNQCRAGRDDRLNRFFDRVGADYYVWE
SQGEAALAGVVGAIVDGDVPTEVPNVFLRSATGEWKLTRKRPEANDLNACSVRWNRFDPT
VLGPTLSTRTARSCAFSCAFCDYPERAGALTLADLSTVERELEELADMGVKRVAFIDDTF
NVPMRRFKDLCRMLVRRDFGIEWFSYFRCGHAREPDVYDLMYDSGCRGVLLGIESGDDQV
LLNMDKRATTAHYQYGIEQLKARGIFTHASFVIGFPGESPQTVQNTIDFINRAGPDTFAV
NHWYYMHATPIHVRAPQFELTGNGHTWSHRSMNSREALEAADQIFDSVVNAAWMPVNGLD
FWGVPYLLGKGMDRSEIVRFLELAKPLTVANVSRGSAEEAAAAEHRFREFVTGLDLAPAR
YRTAGSEFR
Conserved catalytic residues (as determined by automated alignment to family, subgroup, or superfamily HMMs) are shown with teal highlighting . Conserved catalytic residues which do not matched the Conserved Alignment Residue are shown with maroon highlighting . Information regarding their function can be found in the Conserved Residues section below.