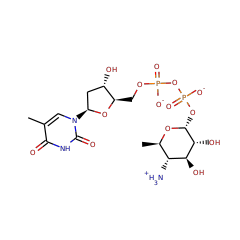
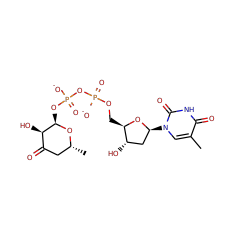
This family of enzymes facilitates the redox-neutral C4 deamination of TDP-4-amino-4,6-dideoxy-D-glucose (1) to generate TDP-4,6-dideoxy-3-keto-D-glucose. Similar to B12-dependent ethanolamine ammonia lyase
DesII can also accept TDP-D-quinovose, in which the C4 amino group of 1 is replaced with a hydroxy group, as a substrate. However, DesII does not catalyze elimination of the C4 hydroxy group, but rather catalyzes oxidation of the C3 hydroxy group. This second, dehydrogenase activity of DesII is analogous to the oxidation of 3-deoxy-scyllo-inosamine by the radical SAM enzyme BtrN from the butirosin biosynthetic pathway8 and the dehydrogenation of a cysteine or serine residue to formylglycine catalyzed by anaerobic sulfatase maturating enzymes
Ruszczycky MW, Ogasawara Y, Liu HW
Radical SAM enzymes in the biosynthesis of sugar-containing natural products
▸ Abstract
Carbohydrates play a key role in the biological activity of numerous natural products. In many instances their biosynthesis requires radical mediated rearrangements, some of which are catalyzed by radical SAM enzymes. BtrN is one such enzyme responsible for the dehydrogenation of a secondary alcohol in the biosynthesis of 2-deoxystreptamine. DesII is another example that catalyzes a deamination reaction necessary for the net C4 deoxygenation of a glucose derivative en route to desosamine formation. BtrN and DesII represent the two most extensively characterized radical SAM enzymes involved in carbohydrate biosynthesis. In this review, we summarize the biosynthetic roles of these two enzymes, their mechanisms of catalysis, the questions that have arisen during these investigations and the insight they can offer for furthering our understanding of radical SAM enzymology. This article is part of a Special Issue entitled: Radical SAM enzymes and Radical Enzymology.
Biochim Biophys Acta
2012;1824(11):1231-1244
| PubMed ID:
22172915
Xue Y, Zhao L, Liu HW, Sherman DH
A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity
▸ Abstract
In a survey of microbial systems capable of generating unusual metabolite structural variability, Streptomyces venezuelae ATCC 15439 is notable in its ability to produce two distinct groups of macrolide antibiotics. Methymycin and neomethymycin are derived from the 12-membered ring macrolactone 10-deoxymethynolide, whereas narbomycin and pikromycin are derived from the 14-membered ring macrolactone, narbonolide. This report describes the cloning and characterization of the biosynthetic gene cluster for these antibiotics. Central to the cluster is a polyketide synthase locus (pikA) that encodes a six-module system comprised of four multifunctional proteins, in addition to a type II thioesterase (TEII). Immediately downstream is a set of genes for desosamine biosynthesis (des) and macrolide ring hydroxylation. The study suggests that Pik TEII plays a role in forming a metabolic branch through which polyketides of different chain length are generated, and the glycosyl transferase (encoded by desVII) has the ability to catalyze glycosylation of both the 12- and 14-membered ring macrolactones. Moreover, the pikC-encoded P450 hydroxylase provides yet another layer of structural variability by introducing regiochemical diversity into the macrolide ring systems. The data support the notion that the architecture of the pik gene cluster as well as the unusual substrate specificity of particular enzymes contributes to its ability to generate four macrolide antibiotics.
Proc Natl Acad Sci U S A
1998;95(21):12111-12116
| PubMed ID:
9770448
Ruszczycky MW, Choi SH, Mansoorabadi SO, Liu HW
Mechanistic studies of the radical S-adenosyl-L-methionine enzyme DesII: EPR characterization of a radical intermediate generated during its catalyzed dehydrogenation of TDP-D-quinovose
▸ Abstract
DesII, a radical S-adenosyl-l-methionine (SAM) enzyme from Streptomyces venezuelae, catalyzes the deamination of TDP-4-amino-4,6-dideoxy-D-glucose to TDP-3-keto-4,6-dideoxy-D-glucose in the desosamine biosynthetic pathway. DesII can also catalyze the dehydrogenation of TDP-D-quinovose to the corresponding 3-keto sugar. Similar to other radical SAM enzymes, DesII catalysis has been proposed to proceed via a radical mechanism. This hypothesis is now confirmed by EPR spectroscopy with the detection of a TDP-D-quinovose radical intermediate having a g-value of 2.0025 with hyperfine coupling to two spin 1/2 nuclei, each with a splitting constant of 33.6 G. A significant decrease in the EPR line width is observed when the radical is generated in reactions conducted in D(2)O versus H(2)O. These results are consistent with a C3 α-hydroxyalkyl radical in which the p-orbital harboring the unpaired electron spin at C3 is periplanar with the C-H bonds at both C2 and C4.
J Am Chem Soc
2011;133(19):7292-7295
| PubMed ID:
21513273
Ko Y, Ruszczycky MW, Choi SH, Liu HW
Mechanistic Studies of the Radical S-Adenosylmethionine Enzyme DesII with TDP-D-Fucose
▸ Abstract
DesII is a radical S-adenosylmethionine (SAM) enzyme that catalyzes the C4-deamination of TDP-4-amino-4,6-dideoxyglucose through a C3 radical intermediate. However, if the C4 amino group is replaced with a hydroxy group (to give TDP-quinovose), the hydroxy group at C3 is oxidized to a ketone with no C4-dehydration. It is hypothesized that hyperconjugation between the C4 CN/O bond and the partially filled p orbital at C3 of the radical intermediate modulates the degree to which elimination competes with dehydrogenation. To investigate this hypothesis, the reaction of DesII with the C4-epimer of TDP-quinovose (TDP-fucose) was examined. The reaction primarily results in the formation of TDP-6-deoxygulose and likely regeneration of TDP-fucose. The remainder of the substrate radical partitions roughly equally between C3-dehydrogenation and C4-dehydration. Thus, changing the stereochemistry at C4 permits a more balanced competition between elimination and dehydrogenation.
Angew Chem Int Ed Engl
2014;None(None):None-None
| PubMed ID:
25418063
Lin GM, Choi SH, Ruszczycky MW, Liu HW
Mechanistic Investigation of the Radical S-Adenosyl-l-methionine Enzyme DesII Using Fluorinated Analogues
▸ Abstract
DesII is a radical S-adenosyl-l-methionine (SAM) enzyme that can act as a deaminase or a dehydrogenase depending on the nature of its TDP-sugar substrate. Previous work has implicated a substrate-derived, C3-centered α-hydroxyalkyl radical as a key intermediate during catalysis. Although deprotonation of the α-hydroxyalkyl radical has been shown to be important for dehydrogenation, much less is known regarding the course of the deamination reaction. To investigate the role played by the C3 hydroxyl during deamination, 3-deutero-3-fluoro analogues of both substrates were prepared and characterized with DesII. In neither case was deamination or oxidation observed; however, in both cases deuterium was efficiently exchanged between the substrate analogues and SAM. These results imply that the C3 hydroxyl plays a key role in both reactions-thereby arguing against a 1,2-migration mechanism of deamination-and that homolysis of SAM concomitant with H atom abstraction from the substrate is readily reversible when forward partitioning is inhibited.
J Am Chem Soc
2015;None(None):None-None
| PubMed ID:
25826575