Top Level Name
⌊ Superfamily (core) Radical SAM
⌊ Subgroup B12-binding domain containing
⌊ Family fortimicin-like methyltransferase
| Total |
100%  |
<100%  |
|||
| Functional domains | 3 | 3 | 0 | ||
| UniProtKB | 6 | 6 | 0 | ||
| GI | 8 | 8 | 0 | ||
| Structures | 0 | ||||
| Reactions | 1 | ||||
| Functional domains of this family were last updated on June 10, 2017 | |||||
| New functional domains were last added to this family on July 25, 2012 | |||||
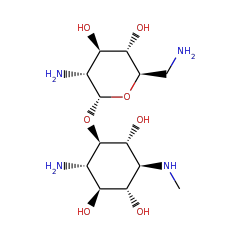
Responsible for the B12 dependent methylation of fortimicin.
Kuzuyama T, Seki T, Dairi T, Hidaka T, Seto H
Nucleotide sequence of fortimicin KL1 methyltransferase gene isolated from Micromonospora olivasterospora, and comparison of its deduced amino acid sequence with those of methyltransferases involved in the biosynthesis of bialaphos and fosfomycin
▸ Abstract
J Antibiot (Tokyo) 1995;48(10):1191-1193 | PubMed ID: 7490235
No notes.
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| network.fam416.bs60.mek250K.xgmml | One node per sequence network | min bit score = 60 max edge count = 250K |
size = 23K num_edges = 3 num_nodes = 3 |
| sfld_alignment_fam416.msa | Annotated Sequence Alignment, Stockholm format | 3 sequences size: 3.8K |