Chandra T, Silver SC, Zilinskas E, Shepard EM, Broderick WE, Broderick JB
Spore photoproduct lyase catalyzes specific repair of the 5R but not the 5S spore photoproduct
▸ Abstract
Bacterial spores are remarkable in their resistance to chemical and physical stresses, including exposure to UV radiation. The unusual UV resistance of bacterial spores is a result of the unique photochemistry of spore DNA, which results in accumulation of 5-thyminyl-5,6-dihydrothymine (spore photoproduct, or SP), coupled with the efficient repair of accumulated damage by the enzyme spore photoproduct lyase (SPL). SPL is a member of the radical AdoMet superfamily of enzymes, and utilizes an iron-sulfur cluster and S-adenosylmethionine to repair SP by a direct reversal mechanism initiated by H atom abstraction from C-6 of the thymine dimer. While two distinct diastereomers of SP (5R or 5S) could in principle be formed upon UV irradiation of bacterial spores, only the 5R configuration is possible for SP formed from adjacent thymines in double helical DNA, due to the constraints imposed by the DNA structure; the 5S configuration is possible in less well-defined DNA structures or as an interstrand cross-link. We report here results from HPLC and MS analysis of in vitro enzymatic assays on stereochemically defined SP substrates demonstrating that SPL specifically repairs only the 5R isomer of SP. The observation that 5R-SP, but not 5S-SP, is a substrate for SPL is consistent with the expectation that 5R is the SP isomer produced in vivo upon UV irradiation of bacterial spore DNA.
J Am Chem Soc
2009;131(7):2420-2421
| PubMed ID:
19178276
Silver SC, Chandra T, Zilinskas E, Ghose S, Broderick WE, Broderick JB
Complete stereospecific repair of a synthetic dinucleotide spore photoproduct by spore photoproduct lyase
▸ Abstract
Spore photoproduct lyase (SP lyase), a member of the radical S-adenosylmethionine superfamily of enzymes, catalyzes the repair of 5-thyminyl-5,6-dihydrothymine [spore photoproduct (SP)], a type of UV-induced DNA damage unique to bacterial spores. The anaerobic purification and characterization of Clostridium acetobutylicum SP lyase heterologously expressed in Escherichia coli, and its catalytic activity in repairing stereochemically defined synthetic dinucleotide SPs was investigated. The purified enzyme contains between 2.3 and 3.1 iron atoms per protein. Electron paramagnetic resonance (EPR) spectroscopy reveals an isotropic signal centered at g = 1.99, characteristic of a [3Fe-4S](+) cluster accounting for 3-4% of the iron in the sample. Upon reduction, a nearly axial signal (g = 2.03, 1.93 and 1.92) characteristic of a [4Fe-4S](+) cluster is observed that accounts for 34-45% of total iron. Addition of S-adenosylmethionine to the reduced enzyme produces a rhombic signal (g = 2.02, 1.93, 1.82) unique to the S-adenosyl-L: -methionine complex while decreasing the overall EPR intensity. This reduced enzyme is shown to rapidly and completely repair the 5R diastereomer of a synthetic dinucleotide SP with a specific activity of 7.1 +/- 0.6 nmol min(-1) mg(-1), whereas no repair was observed for the 5S diastereomer.
J Biol Inorg Chem
2010;15(6):943-955
| PubMed ID:
20405152
Pieck J.C., Hennecke U., Pierik A.J., Friedel M.G., Carell T.
Characterization of a new thermophilic spore photoproduct lyase from Geobacillus stearothermophilus (SplG) with defined lesion containing DNA substrates
▸ Abstract
The Geobacillus stearothermophilus splG gene encodes a thermophilic spore photoproduct lyase (SplG) that belongs to the family of radical S-adenosylmethionine (AdoMet) enzymes. The aerobically purified apo-SplG forms a homodimer, which contains one [4Fe-4S] cluster per monomer unit after reconstitution to the holoform. Formation of the [4Fe-4S] cluster was proven by quantification of the amount of iron and sulfur per homodimer and by UV and EPR spectroscopy. The UV spectrum features a characteristic absorbance at 420 nm typical for [4Fe-4S] clusters, and the EPR data were found to be identical to those of other proteins containing an [4Fe-4S]+ center. Probing of the activity of the holo-SplG with oligonucleotides containing one spore photoproduct lesion at a defined site proved that the enzyme is able to turn over substrate. In addition to repair, we observed cleavage of AdoMet to generate 5'-deoxyadenosine. In the presence of aza-AdoMet the SplG is completely inhibited, which provides direct support for the repair mechanism.
J. Biol. Chem.
2006;281(47):36317-36326
| PubMed ID:
16968710
Friedel MG, Berteau O, Pieck JC, Atta M, Ollagnier-de-Choudens S, Fontecave M, Carell T
The spore photoproduct lyase repairs the 5S- and not the 5R-configured spore photoproduct DNA lesion
▸ Abstract
The spore photoproduct lyase is a Fe-S/AdoMet DNA repair enzyme, which directly repairs spore lesions, induced by UV irradiation of spores, using an unknown radical mechanism. The air sensitive radical SAM enzyme was for the first time challenged with synthetically pure substrates. It was found that the enzyme recognizes a synthetic 5S-configured spore lesion without the central phosphodiester bond. The 5R-configured lesion is in contrast to current belief not a substrate.
Chem Commun (Camb)
2006;28(4):445-447
| PubMed ID:
16493831
Buis JM, Cheek J, Kalliri E, Broderick JB
Characterization of an active spore photoproduct lyase, a DNA repair enzyme in the radical S-adenosylmethionine superfamily
▸ Abstract
The major photoproduct in UV-irradiated Bacillus spore DNA is a unique thymine dimer called spore photoproduct (SP, 5-thyminyl-5,6-dihydrothymine). The enzyme spore photoproduct lyase (SP lyase) has been found to catalyze the repair of SP dimers to thymine monomers in a reaction that requires S-adenosylmethionine. We present here the first detailed characterization of catalytically active SP lyase, which has been anaerobically purified from overexpressing Escherichia coli. Anaerobically purified SP lyase is monomeric and is red-brown in color. The purified enzyme contains approximately 3.1 iron and 3.0 acid-labile S(2-) per protein and has a UV-visible spectrum characteristic of iron-sulfur proteins (410 nm (11.9 mM(-1) cm(-1)) and 450 nm (10.5 mM(-1) cm(-1))). The X-band EPR spectrum of the purified enzyme shows a nearly isotropic signal (g = 2.02) characteristic of a [3Fe-4S]1+ cluster; reduction of SP lyase with dithionite results in the appearance of a new EPR signal (g = 2.03, 1.93, and 1.89) with temperature dependence and g values consistent with its assignment to a [4Fe-4S]1+ cluster. The reduced purified enzyme is active in SP repair, with a specific activity of 0.33 micromol/min/mg. Only a catalytic amount of S-adenosylmethionine is required for DNA repair, and no irreversible cleavage of S-adenosylmethionine into methionine and 5'-deoxyadenosine is observed during the reaction. Label transfer from [5'-3H]S-adenosylmethionine to repaired thymine is observed, providing evidence to support a mechanism in which a 5'-deoxyadenosyl radical intermediate directly abstracts a hydrogen from SP C-6 to generate a substrate radical, and subsequent to radical-mediated beta-scission, a product thymine radical abstracts a hydrogen from 5'-deoxyadenosine to regenerate the 5'-deoxyadenosyl radical. Together, our results support a mechanism in which S-adenosylmethionine acts as a catalytic cofactor, not a substrate, in the DNA repair reaction.
J Biol Chem
2006;281(36):25994-26003
| PubMed ID:
16829680
Chandor A, Berteau O, Douki T, Gasparutto D, Sanakis Y, Ollagnier-de-Choudens S, Atta M, Fontecave M
Dinucleotide spore photoproduct, a minimal substrate of the DNA repair spore photoproduct lyase enzyme from Bacillus subtilis
▸ Abstract
The overwhelming majority of DNA photoproducts in UV-irradiated spores is a unique thymine dimer called spore photoproduct (SP, 5-thymine-5,6-dihydrothymine). This lesion is repaired by the spore photoproduct lyase (SP lyase) enzyme that directly reverts SP to two unmodified thymines. The SP lyase is an S-adenosylmethionine-dependent iron-sulfur protein that belongs to the radical S-adenosylmethionine superfamily. In this study, by using a well characterized preparation of the SP lyase enzyme from Bacillus subtilis, we show that SP in the form of a dinucleoside monophosphate (spore photoproduct of thymidilyl-(3'-5')-thymidine) is efficiently repaired, allowing a kinetic characterization of the enzyme. The preparation of this new substrate is described, and its identity is confirmed by mass spectrometry and comparison with authentic spore photoproduct. The fact that the spore photoproduct of thymidilyl-(3'-5')-thymidine dimer is repaired by SP lyase may indicate that the SP lesion does not absolutely need to be contained within a single- or double-stranded DNA for recognition and repaired by the SP lyase enzyme.
J Biol Chem
2006;281(37):26922-26931
| PubMed ID:
16829676
Benjdia A, Heil K, Barends TR, Carell T, Schlichting I
Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme
▸ Abstract
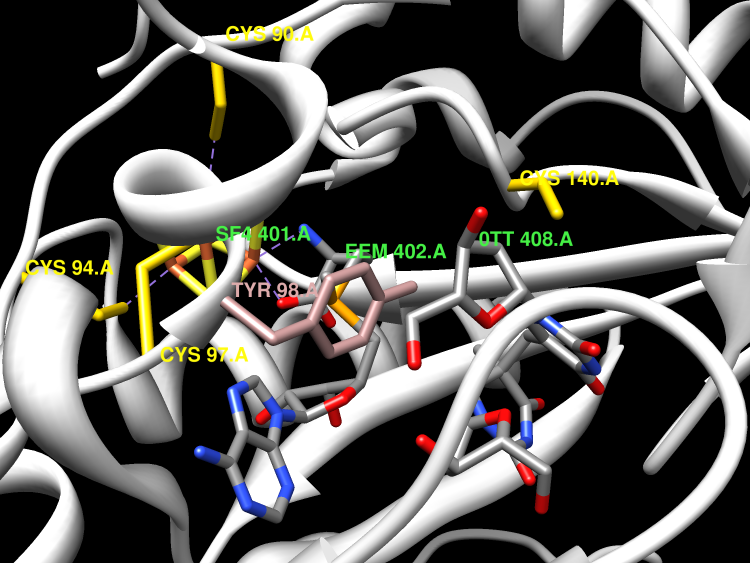
Bacterial spores possess an enormous resistance to ultraviolet (UV) radiation. This is largely due to a unique DNA repair enzyme, Spore Photoproduct Lyase (SP lyase) that repairs a specific UV-induced DNA lesion, the spore photoproduct (SP), through an unprecedented radical-based mechanism. Unlike DNA photolyases, SP lyase belongs to the emerging superfamily of radical S-adenosyl-l-methionine (SAM) enzymes and uses a [4Fe-4S](1+) cluster and SAM to initiate the repair reaction. We report here the first crystal structure of this enigmatic enzyme in complex with its [4Fe-4S] cluster and its SAM cofactor, in the absence and presence of a DNA lesion, the dinucleoside SP. The high resolution structures provide fundamental insights into the active site, the DNA lesion recognition and binding which involve a β-hairpin structure. We show that SAM and a conserved cysteine residue are perfectly positioned in the active site for hydrogen atom abstraction from the dihydrothymine residue of the lesion and donation to the α-thyminyl radical moiety, respectively. Based on structural and biochemical characterizations of mutant proteins, we substantiate the role of this cysteine in the enzymatic mechanism. Our structure reveals how SP lyase combines specific features of radical SAM and DNA repair enzymes to enable a complex radical-based repair reaction to take place.
Nucleic Acids Res
2012;None(None):None-None
| PubMed ID:
22761404
Yang L, Li L
The enzyme-mediated direct reversal of a dithymine photoproduct in germinating endospores
▸ Abstract
Spore photoproduct lyase (SPL) repairs a special thymine dimer, 5-thyminyl-5,6-dihydrothymine, which is commonly called spore photoproduct, or SP, in germinating endospores. SP is the exclusive DNA photo-damaging product found in endospores; its generation and swift repair by SPL are responsible for the spores' extremely high UV resistance. Early in vivo studies suggested that SPL utilizes a direct reversal strategy to repair SP in the absence of light. Recently, it has been established that SPL belongs to the radical S-adenosylmethionine (SAM) superfamily. The enzymes in this superfamily utilize a tri-cysteine CXXXCXXC motif to bind a [4Fe-4S] cluster. The cluster provides an electron to the S-adenosylmethionine (SAM) to reductively cleave its C5'-S bond, generating a reactive 5'-deoxyadenosyl (5'-dA) radical. This 5'-dA radical abstracts the proR hydrogen atom from the C6 carbon of SP to initiate the repair process; the resulting SP radical subsequently fragments to generate a putative thymine methyl radical, which accepts a back-donated H atom to yield the repaired TpT. The H atom donor is suggested to be a conserved cysteine141 in B. subtilis SPL; the resulting thiyl radical likely interacts with a neighboring tyrosine99 before oxidizing the 5'-dA to 5'-dA radical and, subsequently, regenerating SAM. These findings suggest SPL to be the first enzyme in the large radical SAM superfamily (>44,000 members) to utilize a radical transfer pathway for catalysis; its study should shed light on the mechanistic understanding of the SAM regeneration process in other members of the superfamily.
Int J Mol Sci
2013;14(7):13137-13153
| PubMed ID:
23799365
Silver SC, Gardenghi DJ, Naik SG, Shepard EM, Huynh BH, Szilagyi RK, Broderick JB
Combined Mössbauer spectroscopic, multi-edge X-ray absorption spectroscopic, and density functional theoretical study of the radical SAM enzyme spore photoproduct lyase
▸ Abstract
Spore photoproduct lyase (SPL), a member of the radical S-adenosyl-L-methionine (SAM) superfamily, catalyzes the direct reversal of the spore photoproduct, a thymine dimer specific to bacterial spores, to two thymines. SPL requires SAM and a redox-active [4Fe-4S] cluster for catalysis. Mössbauer analysis of anaerobically purified SPL indicates the presence of a mixture of cluster states with the majority (40 %) as [2Fe-2S](2+) clusters and a smaller amount (15 %) as [4Fe-4S](2+) clusters. On reduction, the cluster content changes to primarily (60 %) [4Fe-4S](+). The speciation information from Mössbauer data allowed us to deconvolute iron and sulfur K-edge X-ray absorption spectra to uncover electronic (X-ray absorption near-edge structure, XANES) and geometric (extended X-ray absorption fine structure, EXAFS) structural features of the Fe-S clusters, and their interactions with SAM. The iron K-edge EXAFS data provide evidence for elongation of a [2Fe-2S] rhomb of the [4Fe-4S] cluster on binding SAM on the basis of an Fe···Fe scatterer at 3.0 Å. The XANES spectra of reduced SPL in the absence and presence of SAM overlay one another, indicating that SAM is not undergoing reductive cleavage. The X-ray absorption spectroscopy data for SPL samples and data for model complexes from the literature allowed the deconvolution of contributions from [2Fe-2S] and [4Fe-4S] clusters to the sulfur K-edge XANES spectra. The analysis of pre-edge features revealed electronic changes in the Fe-S clusters as a function of the presence of SAM. The spectroscopic findings were further corroborated by density functional theory calculations that provided insights into structural and electronic perturbations that can be correlated by considering the role of SAM as a catalyst or substrate.
J Biol Inorg Chem
2014;19(3):465-483
| PubMed ID:
24532333
Benjdia A, Heil K, Winkler A, Carell T, Schlichting I
Rescuing DNA repair activity by rewiring the H-atom transfer pathway in the radical SAM enzyme, spore photoproduct lyase
▸ Abstract
The radical SAM enzyme, spore photoproduct lyase, requires an H-atom transfer (HAT) pathway to catalyze DNA repair. By rational engineering, we demonstrate that it is possible to rewire its HAT pathway, a first step toward the development of novel catalysts based on the radical SAM enzyme scaffold.
Chem Commun (Camb)
2014;50(91):14201-14204
| PubMed ID:
25285338
Ghose S, Hilmer JK, Bothner B, Broderick JB
Solution phase dynamics of the DNA repair enzyme spore photoproduct lyase as probed by H/D exchange
▸ Abstract
Spore photoproduct lyase (SPL) catalyzes the repair of the UV lesion spore photoproduct (SP) in a reaction dependent on S-adenosyl-L-methionine (SAM). We have utilized H/D exchange to show that in the presence of SAM, a significant reduction in H/D exchange is observed upon binding SPTpT or undamaged oligonucleotide, indicating a shift of 20 or 10 amide protons, respectively, from a rapidly-exchangable state to a fully-protected conformation. In the absence of SAM, neither the oligonucleotide nor the SPTpT produce a significant perturbation in H/D exchange, indicating SAM is a requisite binding partner. Performing the same experiments in aerobic conditions reduced the magnitude of ligand-induced structural changes, consistent with the importance of the oxygen-sensitive iron-sulfur cluster for SAM and substrate binding.
FEBS Lett
2014;588(17):3023-3029
| PubMed ID:
24950428