Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE Jr, Marletta MA
Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein
▸ Abstract
The Escherichia coli lipA gene product has been genetically linked to carbon-sulfur bond formation in lipoic acid biosynthesis [Vanden Boom, T. J., Reed, K. E., and Cronan, J. E., Jr. (1991) J. Bacteriol. 173, 6411-6420], although in vitro lipoate biosynthesis with LipA has never been observed. In this study, the lipA gene and a hexahistidine tagged lipA construct (LipA-His) were overexpressed in E. coli as soluble proteins. The proteins were purified as a mixture of monomeric and dimeric species that contain approximately four iron atoms per LipA polypeptide and a similar amount of acid-labile sulfide. Electron paramagnetic resonance and electronic absorbance spectroscopy indicate that the proteins contain a mixture of [3Fe-4S] and [4Fe-4S] cluster states. Reduction with sodium dithionite results in small quantities of an S = 1/2 [4Fe-4S](1+) cluster with the majority of the protein containing a species consistent with an S = 0 [4Fe-4S](2+) cluster. LipA was assayed for lipoate or lipoyl-ACP formation using E. coli lipoate-protein ligase A (LplA) or lipoyl-[acyl-carrier-protein]-protein-N-lipoyltransferase (LipB), respectively, to lipoylate apo-pyruvate dehydrogenase complex (apo-PDC) [Jordan, S. W., and Cronan, J. E. (1997) Methods Enzymol. 279, 176-183]. When sodium dithionite-reduced LipA was incubated with octanoyl-ACP, LipB, apo-PDC, and S-adenosyl methionine (AdoMet), lipoylated PDC was formed. As shown by this assay, octanoic acid is not a substrate for LipA. Confirmation that LipA catalyzes formation of lipoyl groups from octanoyl-ACP was obtained by MALDI mass spectrometry of a recombinant PDC lipoyl-binding domain that had been lipoylated in a LipA reaction. These results provide information about the mechanism of LipA catalysis and place LipA within the family of iron-sulfur proteins that utilize AdoMet for radical-based chemistry.
Biochemistry
2000;39(49):15166-15178
| PubMed ID:
11106496
Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE
Assembly of the covalent linkage between lipoic acid and its cognate enzymes
▸ Abstract
Lipoic acid is synthesized from octanoic acid by insertion of sulfur atoms at carbons 6 and 8 and is covalently attached to a pyruvate dehydrogenase (PDH) subunit. We show that sulfur atoms can be inserted into octanoyl moieties attached to a PDH subunit or a derived domain. Escherichia coli lipB mutants grew well when supplemented with octanoate in place of lipoate. Octanoate growth required both lipoate protein ligase (LplA) and LipA, the sulfur insertion protein, suggesting that LplA attached octanoate to the dehydrogenase and LipA then converted the octanoate to lipoate. This pathway was tested by labeling a PDH domain with deuterated octanoate in an E. coli strain devoid of LipA activity. The labeled octanoyl domain was converted to lipoylated domain upon restoration of LipA. Moreover, octanoyl domain and octanoyl-PDH were substrates for sulfur insertion in vitro.
Chem Biol.
2003;10(12):1293-1302
| PubMed ID:
14700636
Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ
Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid
▸ Abstract
Lipoyl synthase (LipA) catalyzes the formation of the lipoyl cofactor, which is employed by several multienzyme complexes for the oxidative decarboxylation of various alpha-keto acids, as well as the cleavage of glycine into CO(2) and NH(3), with concomitant transfer of its alpha-carbon to tetrahydrofolate, generating N(5),N(10)-methylenetetrahydrofolate. In each case, the lipoyl cofactor is tethered covalently in an amide linkage to a conserved lysine residue located on a designated lipoyl-bearing subunit of the complex. Genetic and biochemical studies suggest that lipoyl synthase is a member of a newly established class of metalloenzymes that use S-adenosyl-l-methionine (AdoMet) as a source of a 5'-deoxyadenosyl radical (5'-dA(*)), which is an obligate intermediate in each reaction. These enzymes contain iron-sulfur clusters, which provide an electron during the cleavage of AdoMet, forming l-methionine in addition to the primary radical. Recently, one substrate for lipoyl synthase has been shown to be the octanoylated derivative of the lipoyl-bearing subunit (E(2)) of the pyruvate dehydrogenase complex [Zhao, S., Miller, J. R., Jian, Y., Marletta, M. A., and Cronan, J. E., Jr. (2003) Chem. Biol. 10, 1293-1302]. Herein, we show that the octanoylated derivative of the lipoyl-bearing subunit of the glycine cleavage system (H-protein) is also a substrate for LipA, providing further evidence that the cofactor is synthesized on its target protein. Moreover, we show that the 5'-dA(*) acts directly on the octanoyl substrate, as evidenced by deuterium transfer from [octanoyl-d(15)]H-protein to 5'-deoxyadenosine. Last, our data indicate that 2 equiv of AdoMet are cleaved irreversibly in forming 1 equiv of [lipoyl]H-protein and are consistent with a model in which two LipA proteins are required to synthesize one lipoyl group.
Biochemistry
2004;43(21):6378-6386
| PubMed ID:
15157071
Cicchillo RM, Lee KH, Baleanu-Gogonea C, Nesbitt NM, Krebs C, Booker SJ.
Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide.
▸ Abstract
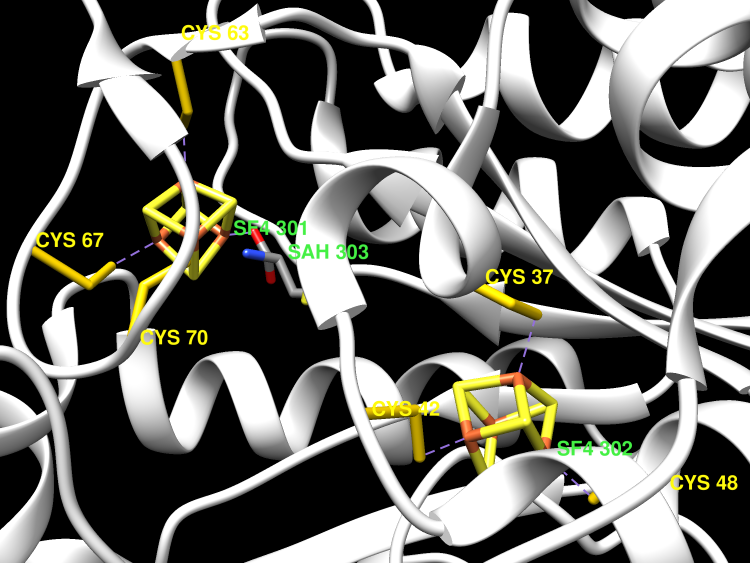
Lipoyl synthase (LS) is a member of a recently established class of metalloenzymes that use S-adenosyl-l-methionine (SAM) as the precursor to a high-energy 5'-deoxyadenosyl 5'-radical (5'-dA(*)). In the LS reaction, the 5'-dA(*) is hypothesized to abstract hydrogen atoms from C-6 and C-8 of protein-bound octanoic acid with subsequent sulfur insertion, generating the lipoyl cofactor. Consistent with this premise, 2 equiv of SAM is required to synthesize 1 equiv of the lipoyl cofactor, and deuterium transfer from octanoyl-d(15) H-protein of the glycine cleavage system-one of the substrates for LS-has been reported [Cicchillo, R. M., Iwig, D. F., Jones, A. D., Nesbitt, N. M., Baleanu-Gogonea, C., Souder, M. G., Tu, L., and Booker, S. J. (2004) Biochemistry 43, 6378-6386]. However, the exact identity of the sulfur donor remains unknown. We report herein that LS from Escherichia coli can accommodate two [4Fe-4S] clusters per polypeptide and that this form of the enzyme is relevant to turnover. One cluster is ligated by the cysteine amino acids in the C-X(3)-C-X(2)-C motif that is common to all radical SAM enzymes, while the other is ligated by the cysteine amino acids residing in a C-X(4)-C-X(5)-C motif, which is conserved only in lipoyl synthases. When expressed in the presence of a plasmid that harbors an Azotobacter vinelandii isc operon, which is involved in Fe/S cluster biosynthesis, the as-isolated wild-type enzyme contained 6.9 +/- 0.5 irons and 6.4 +/- 0.9 sulfides per polypeptide and catalyzed formation of 0.60 equiv of 5'-deoxyadenosine (5'-dA) and 0.27 equiv of lipoylated H-protein per polypeptide. The C68A-C73A-C79A triple variant, expressed and isolated under identical conditions, contained 3.0 +/- 0.1 irons and 3.6 +/- 0.4 sulfides per polypeptide, while the C94A-C98A-C101A triple variant contained 4.2 +/- 0.1 irons and 4.7 +/- 0.8 sulfides per polypeptide. Neither of these variant proteins catalyzed formation of 5'-dA or the lipoyl group. Mössbauer spectroscopy of the as-isolated wild-type protein and the two triple variants indicates that greater than 90% of all associated iron is in the configuration [4Fe-4S](2+). When wild-type LS was reconstituted with (57)Fe and sodium sulfide, it harbored considerably more iron (13.8 +/- 0.6) and sulfide (13.1 +/- 0.2) per polypeptide and catalyzed formation of 0.96 equiv of 5'-dA and 0.36 equiv of the lipoyl group. Mössbauer spectroscopy of this protein revealed that only approximately 67% +/- 6% of the iron is in the form of [4Fe-4S](2+) clusters, amounting to 9.2 +/- 0.4 irons and 8.8 +/- 0.1 sulfides or 2 [4Fe-4S](2+) clusters per polypeptide, with the remainder of the iron occurring as adventitiously bound species. Although the Mössbauer parameters of the clusters associated with each of the variants are similar, EPR spectra of the reduced forms of the cluster show small differences in spin concentration and g-values, consistent with each of these clusters as distinct species residing in each of the two cysteine-containing motifs.
Biochemistry
2004;43(37):11770-11781
| PubMed ID:
15362861
Cicchillo RM, Booker SJ.
Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide.
▸ Abstract
Lipoyl synthase catalyzes the final step in the de novo biosynthesis of the lipoyl cofactor, which is the insertion of two sulfur atoms into an octanoyl chain that is bound in an amide linkage to a conserved lysine on a lipoyl-accepting protein. We show herein that the sulfur atoms in the lipoyl cofactor are derived from lipoyl synthase itself, and that each lipoyl synthase polypeptide contributes both of the sulfur atoms to the intact cofactor.
J Am Chem Soc.
2005;127(9):2860-2861
| PubMed ID:
15740115
Booker SJ
Unraveling the pathway of lipoic acid biosynthesis
▸ Abstract
Lipoic acid is almost universally required for aerobic metabolism. However, the mechanism for its synthesis and incorporation into proteins has remained elusive. A groundbreaking study published in the December issue of Chemistry & Biology uncovers critical features of the lipoic acid biosynthetic pathway.
Comment on
Assembly of the covalent linkage between lipoic acid and its cognate enzymes. [Chem Biol. 2003]
Chem Biol.
2004;11(1):10-12
| PubMed ID:
15112987
Harmer JE, Hiscox MJ, Dinis PC, Fox SJ, Iliopoulos A, Hussey JE, Sandy J, Van Beek FT, Essex JW, Roach PL
Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions
▸ Abstract
Lipoyl cofactors are essential for living organisms and are produced by the insertion of two sulfur atoms into the relatively unreactive C-H bonds of an octanoyl substrate. This reaction requires lipoyl synthase, a member of the radical SAM enzyme superfamily. Herein we present crystal structures of lipoyl synthase with two [4Fe-4S] clusters bound at opposite ends of the TIM barrel, the usual fold of the radical SAM superfamily. The cluster required for reductive SAM cleavage conserves the features of the radical SAM superfamily, but the auxiliary cluster is bound by a CX4CX5C motif unique to lipoyl synthase. The fourth ligand to the auxiliary cluster is an extremely unusual serine residue. Site directed mutants show this conserved serine ligand is essential for the sulfur insertion steps. One crystallized LipA complex contains MTA, a breakdown product of SAM, bound in the likely SAM binding site. Modelling has identified an 18 Å deep channel, well-proportioned to accommodate an octanoyl substrate. These results suggest the auxiliary cluster is the likely sulfur donor, but access to a sulfide ion for the second sulfur insertion reaction requires the loss of an iron atom from the auxiliary cluster, which the serine ligand may enabled.
Biochem J
2014;None(None):None-None
| PubMed ID:
25100160
Cronan JE
The structure of lipoyl synthase, a remarkable enzyme that performs the last step of an extraordinary biosynthetic pathway
▸ Abstract
Lipoic acid is assembled on its cognate proteins (e.g. the E2 subunit of pyruvate dehydrogenase). An octanoyl moiety is transferred from the octanoyl-ACP of fatty acid synthetase to a specific lysine residue of the cognate protein followed by sulfur insertion at C6 and C8 of the octanoyl chain. The challenging chemistry of this last step is performed by the radical S-adenosylmethionine (SAM) enzyme lipoyl synthase (LipA). In this issue of the Biochemical Journal, Harmer et al. report the first crystal structure of a lipoyl synthase and demonstrate that it contains two [4Fe-4S] clusters, the canonical radical SAM cluster plus a second auxiliary cluster having an unprecedented serine ligand. The structure provides strong support for the model in which the auxiliary cluster donates the lipoate sulfur atoms.
Biochem J
2014;464(1):None-None
| PubMed ID:
25341020
McLaughlin MI, Lanz ND, Goldman PJ, Lee KH, Booker SJ, Drennan CL
Crystallographic snapshots of sulfur insertion by lipoyl synthase
▸ Abstract
Lipoyl synthase (LipA) catalyzes the insertion of two sulfur atoms at the unactivated C6 and C8 positions of a protein-bound octanoyl chain to produce the lipoyl cofactor. To activate its substrate for sulfur insertion, LipA uses a [4Fe-4S] cluster and S-adenosylmethionine (AdoMet) radical chemistry; the remainder of the reaction mechanism, especially the source of the sulfur, has been less clear. One controversial proposal involves the removal of sulfur from a second (auxiliary) [4Fe-4S] cluster on the enzyme, resulting in destruction of the cluster during each round of catalysis. Here, we present two high-resolution crystal structures of LipA from Mycobacterium tuberculosis: one in its resting state and one at an intermediate state during turnover. In the resting state, an auxiliary [4Fe-4S] cluster has an unusual serine ligation to one of the irons. After reaction with an octanoyllysine-containing 8-mer peptide substrate and 1 eq AdoMet, conditions that allow for the first sulfur insertion but not the second insertion, the serine ligand dissociates from the cluster, the iron ion is lost, and a sulfur atom that is still part of the cluster becomes covalently attached to C6 of the octanoyl substrate. This intermediate structure provides a clear picture of iron-sulfur cluster destruction in action, supporting the role of the auxiliary cluster as the sulfur source in the LipA reaction and describing a radical strategy for sulfur incorporation into completely unactivated substrates.
Proc Natl Acad Sci U S A
2016;113(34):9446-9450
| PubMed ID:
27506792
Lanz ND, Rectenwald JM, Wang B, Kakar ES, Laremore TN, Booker SJ, Silakov A
Characterization of a Radical Intermediate in Lipoyl Cofactor Biosynthesis
▸ Abstract
Lipoyl synthase (LipA) catalyzes the final step in the biosynthesis of the lipoyl cofactor, the insertion of two sulfur atoms at C6 and C8 of an n-octanoyl chain. LipA is a member of the radical S-adenosylmethionine (SAM) superfamily of enzymes and uses two [4Fe-4S] clusters to catalyze its transformation. One cluster binds in contact with SAM and donates the requisite electron for the reductive cleavage of SAM to generate two 5'-deoxyadenosyl 5'-radicals, which abstract hydrogen atoms from C6 and C8 of the substrate. By contrast, the second, auxiliary [4Fe-4S] cluster, has been hypothesized to serve as the sulfur donor in the reaction. Such a sacrificial role for an iron-sulfur cluster during catalysis has not been universally accepted. Use of a conjugated 2,4-hexadienoyl-containing substrate analogue has allowed the substrate radical to be trapped and characterized by continuous-wave and pulsed electron paramagnetic resonance methods. Here we report the observation of a (57)Fe hyperfine coupling interaction with the paramagnetic signal, which indicates that the iron-sulfur cluster of LipA and its substrate are within bonding distance.
J Am Chem Soc
2015;137(41):13216-13219
| PubMed ID:
26390103