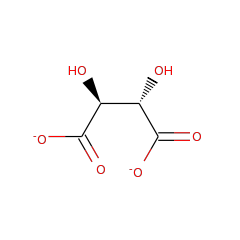
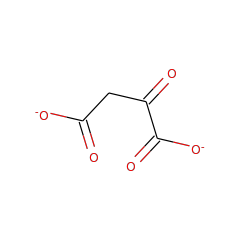
Top Level Name
⌊ Superfamily (core) Enolase
⌊ Subgroup mandelate racemase
⌊ Family D-tartrate dehydratase
⌊ FunctionalDomain D-tartrate dehydratase (ID 397)
No Notes.
| Superfamily Assignment Evidence Code(s) | ISS |
| Family Assignment Evidence Code | CFM PubMed:17144653 |
| This entry was last updated on | Nov. 22, 2017 |
References to Other Databases
Genbank
| Species | GI | Accession | Proteome |
|---|---|---|---|
| Bradyrhizobium japonicum USDA 110 Taxon ID: 224911 | 27381841 | NP_773370.1 (RefSeq) | PRP URP |
| Taxon ID: 374 | 499402002 | WP_011089469.1 (RefSeq) | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920030 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920029 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920028 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920027 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920026 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920025 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920024 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920023 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920022 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920021 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920020 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920019 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920018 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920017 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920016 | URP | |
| Bradyrhizobium japonicum Taxon ID: 375 | 122920015 | URP | |
| Bradyrhizobium japonicum USDA 110 Taxon ID: 224911 | 81735269 | PRP URP | |
| Bradyrhizobium japonicum USDA 110 Taxon ID: 224911 | 27355010 | PRP URP | |
| Show All | |||
Uniprot
| Protein Name | Accession | EC Number
 |
Identifier |
|---|---|---|---|
| D(-)-tartrate dehydratase | Q89FH0 | 4.2.1.81 | TARD_BRADU (Swiss-Prot) |
Length of Enzyme (full-length): 389 | Length of Functional Domain: 387
MSVRIVDVREITKPISSPIRNAYIDFTKMTTSLVAVVTDVVREGKRVVGYGFNSNGRYGQ
GGLIRERFASRILEADPKKLLNEAGDNLDPDKVWAAMMINEKPGGHGERSVAVGTIDMAV
WDAVAKIAGKPLFRLLAERHGVKANPRVFVYAAGGYYYPGKGLSMLRGEMRGYLDRGYNV
VKMKIGGAPIEEDRMRIEAVLEEIGKDAQLAVDANGRFNLETGIAYAKMLRDYPLFWYEE
VGDPLDYALQAALAEFYPGPMATGENLFSHQDARNLLRYGGMRPDRDWLQFDCALSYGLC
EYQRTLEVLKTHGWSPSRCIPHGGHQMSLNIAAGLGLGGNESYPDLFQPYGGFPDGVRVE
NGHITMPDLPGIGFEGKSDLYKEMKALAE
Conserved catalytic residues (as determined by automated alignment to family, subgroup, or superfamily HMMs) are shown with teal highlighting . Conserved catalytic residues which do not matched the Conserved Alignment Residue are shown with maroon highlighting . Information regarding their function can be found in the Conserved Residues section below.