Top Level Name
⌊ Superfamily (core) Radical SAM
⌊ Subgroup SPASM/twitch domain containing
⌊ Family UDP-N-acetyl-tunicamine-uracil synthase (TunB-like)
| Total |
100%  |
<100%  |
|||
| Functional domains | 4 | 4 | 0 | ||
| UniProtKB | 4 | 4 | 0 | ||
| GI | 15 | 15 | 0 | ||
| Structures | 0 | ||||
| Reactions | 1 | ||||
| Functional domains of this family were last updated on June 10, 2017 | |||||
| New functional domains were last added to this family on Aug. 17, 2012 | |||||
Involved in the biosynthesis of tunicamycins. Catalyses the radical coupling reaction in which a 5′-uridyl radical undergoes radical addition at C6 of N-acetyl-galactoseamine-5,6-ene to yield an α-alkoxyalkyl radical. Subsequent hydrogen atom abstraction from 5′-deoxyadenosine could lead to UDP-N-acetyl-tunicamine-uracil and the regeneration of SAM.
Ruszczycky MW, Ogasawara Y, Liu HW
Radical SAM enzymes in the biosynthesis of sugar-containing natural products
▸ Abstract
Biochim Biophys Acta 2012;1824(11):1231-1244 | PubMed ID: 22172915
Whilst this family seems to belong to the SPASM/twitch domain containing subgroup via the similarity network and at the subgroup E-value threshold for the complete superfamily, it does not have a secondary iron-sulfur binding site.
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| network.fam429.bs60.mek250K.xgmml | One node per sequence network | min bit score = 60 max edge count = 250K |
size = 9.4K num_edges = 6 num_nodes = 4 |
| sfld_alignment_fam429.msa | Annotated Sequence Alignment, Stockholm format | 4 sequences size: 2.9K |
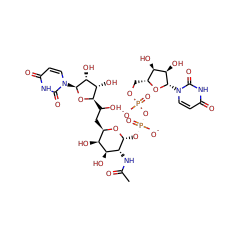
Catalyzed Reaction(s)
UDP-N-acetyl-tunicamine-uracil synthase
 |
+ |  |
 |
 |
||
| uridine 16704 |
UDP-N-acetylgalactosamine-5,6-ene(2-) 85718 |
UDP-N-acetyltunicamine-uracil(2-) 85719 |
EC: 4.-.-.- | IntEnz: 4.-.-.- | Kegg: 4.-.-.- | BioCyc: 4.-.-.- | BRENDA: 4.-.-.- |


