| Total |
100%  |
<100%  |
|||
| Functional domains | 171 | 158 | 13 | ||
| UniProtKB | 181 | 169 | 12 | ||
| GI | 487 | 450 | 37 | ||
| Structures | 0 | ||||
| Reactions | 1 | ||||
| Functional domains of this family were last updated on June 10, 2017 | |||||
| New functional domains were last added to this family on June 22, 2014 | |||||
This family of enzymes is thought to be responsible for the di-methylation of a precursor of methanopterin (MPT). MPT, and it's derivatives, are one-carbon carrier coenzymes that are involved in essential biochemical processes of methanogenesis and methylotrophy performed by specialized archaea and bacteria.
Work on the CFM (MJ0619) demonstrated that this protein is responsible for catalyzing the methylation at both C-7 and C-9 of the 7,8-dihydro-6-hydroxymethylpterin substrate. Site-directed mutagenesis of Cys77 (CFM numbering) present in the first of two canonical radical SAM CX3CX2C motifs present in MJ0619 did not inhibit C-7 methylation while mutation of Cys102, found in the other radical SAM amino acid motif, resulted in loss of C-7 methylation, suggesting that the first motif is likely involved in C-9 methylation while the second motif is required for C-7 methylation. Further experiments demonstrated that the C-7 methyl group is not derived from methionine and methylation does not require cobalamin. This, along with the results of the deuterium labeling experiments suggest that these enzymes utilize a previously uncharacterized mechanism for methylation, using using methylenetetrahydrofolate as a methyl group donor.
Allen KD, Xu H, White RH
Identification of a unique radical SAM methylase likely involved in methanopterin biosynthesis in Methanocaldococcus jannaschii
▸ Abstract
J Bacteriol 2014;None(None):None-None | PubMed ID: 25002541
At an E-value of 1E-75, this group of enzymes appears to be somewhat distinct from the others, and whilst still connected to many other enzymes in this subgroup, in order to be as accurate in function transfer as possible, this set is taken as the family, and the more loosely connected enzymes are considered subgroup only.
Static File Downloads
| File Name | Description | Parameters | Stats |
|---|---|---|---|
| network.fam385.bs60.mek250K.xgmml | One node per sequence network | min bit score = 60 max edge count = 250K |
size = 7.7M num_edges = 14535 num_nodes = 171 |
| sfld_alignment_fam385.msa | Annotated Sequence Alignment, Stockholm format | 92 sequences size: 61K |
Catalyzed Reaction(s)
7,8-dihydro-6-hydroxymethylpterin dimethyltransferase
 |
+ 2 |  |
+ 2 |  |
+ 4 | 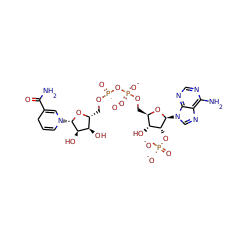 |
 |
 |
+ 2 |  |
+ |  |
+ 2 |  |
+ 4 | 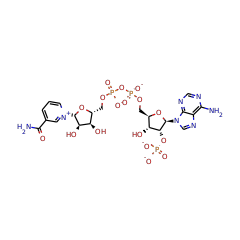 |
||
| 2-amino-6-(hydroxymethyl)-7,8-dihydropteridin-4-one 44841 |
S-adenosyl-L-methioninate 67040 |
(6R)-5,10-methenyltetrahydrofolic acid 15638 |
NADPH(4-) 57783 |
6-hydroxyethyl-7-methylpterin 132441 |
L-methionine zwitterion 57844 |
5'-deoxyadenosine 17319 |
5,6,7,8-tetrahydrofolic acid 20506 |
NADP(3-) 58349 |
EC: | IntEnz: | Kegg: | BioCyc: | BRENDA: |


