Arragain S, Garcia-Serres R, Blondin G, Douki T, Clemancey M, Latour JM, Forouhar F, Neely H, Montelione GT, Hunt JF, Mulliez E, Fontecave M, Atta M
Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase
▸ Abstract
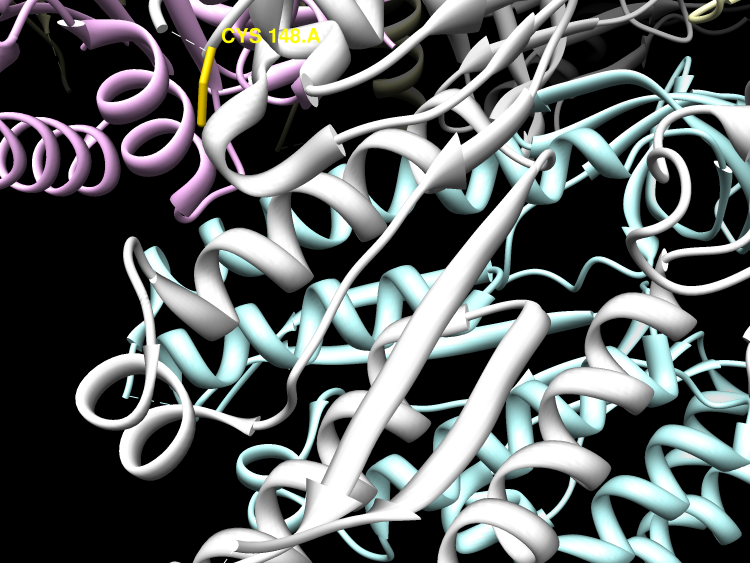
Post-translational modifications of ribosomal proteins are important for the accuracy of the decoding machinery. A recent in vivo study has shown that the rimO gene is involved in generation of the 3-methylthio derivative of residue Asp-89 in ribosomal protein S12 (Anton, B. P., Saleh, L., Benner, J. S., Raleigh, E. A., Kasif, S., and Roberts, R. J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 1826-1831). This reaction is formally identical to that catalyzed by MiaB on the C2 of adenosine 37 near the anticodon of several tRNAs. We present spectroscopic evidence that Thermotoga maritima RimO, like MiaB, contains two [4Fe-4S] centers, one presumably bound to three invariant cysteines in the central radical S-adenosylmethionine (AdoMet) domain and the other to three invariant cysteines in the N-terminal UPF0004 domain. We demonstrate that holo-RimO can specifically methylthiolate the aspartate residue of a 20-mer peptide derived from S12, yielding a mixture of mono- and bismethylthio derivatives. Finally, we present the 2.0 A crystal structure of the central radical AdoMet and the C-terminal TRAM (tRNA methyltransferase 2 and MiaB) domains in apo-RimO. Although the core of the open triose-phosphate isomerase (TIM) barrel of the radical AdoMet domain was conserved, RimO showed differences in domain organization compared with other radical AdoMet enzymes. The unusually acidic TRAM domain, likely to bind the basic S12 protein, is located at the distal edge of the radical AdoMet domain. The basic S12 protein substrate is likely to bind RimO through interactions with both the TRAM domain and the concave surface of the incomplete TIM barrel. These biophysical results provide a foundation for understanding the mechanism of methylthioation by radical AdoMet enzymes in the MiaB/RimO family.
J Biol Chem
2010;285(8):5792-5801
| PubMed ID:
20007320
Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ
Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily
▸ Abstract
RimO, encoded by the yliG gene in Escherichia coli, has been recently identified in vivo as the enzyme responsible for the attachment of a methylthio group on the beta-carbon of Asp88 of the small ribosomal protein S12 [Anton, B. P., Saleh, L., Benner, J. S., Raleigh, E. A., Kasif, S., and Roberts, R. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1826-1831]. To date, it is the only enzyme known to catalyze methylthiolation of a protein substrate; the four other naturally occurring methylthio modifications have been observed on tRNA. All members of the methylthiotransferase (MTTase) family, to which RimO belongs, have been shown to contain the canonical CxxxCxxC motif in their primary structures that is typical of the radical S-adenosylmethionine (SAM) family of proteins. MiaB, the only characterized MTTase, and the enzyme experimentally shown to be responsible for methylthiolation of N(6)-isopentenyladenosine of tRNA in E. coli and Thermotoga maritima, has been demonstrated to harbor two distinct [4Fe-4S] clusters. Herein, we report in vitro biochemical and spectroscopic characterization of RimO. We show by analytical and spectroscopic methods that RimO, overproduced in E. coli in the presence of iron-sulfur cluster biosynthesis proteins from Azotobacter vinelandii, contains one [4Fe-4S](2+) cluster. Reconstitution of this form of RimO (RimO(rcn)) with (57)Fe and sodium sulfide results in a protein that contains two [4Fe-4S](2+) clusters, similar to MiaB. We also show by mass spectrometry that RimO(rcn) catalyzes the attachment of a methylthio group to a peptide substrate analogue that mimics the loop structure bearing aspartyl 88 of the S12 ribosomal protein from E. coli. Kinetic analysis of this reaction shows that the activity of RimO(rcn) in the presence of the substrate analogue does not support a complete turnover. We discuss the possible requirement for an assembled ribosome for fully active RimO in vitro. Our findings are consistent with those of other enzymes that catalyze sulfur insertion, such as biotin synthase, lipoyl synthase, and MiaB.
Biochemistry
2009;48(42):10162-10174
| PubMed ID:
19736993
Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ
RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli
▸ Abstract
Ribosomal protein S12 undergoes a unique posttranslational modification, methylthiolation of residue D88, in Escherichia coli and several other bacteria. Using mass spectrometry, we have identified the enzyme responsible for this modification in E. coli, the yliG gene product. This enzyme, which we propose be called RimO, is a radical-S-adenosylmethionine protein that bears strong sequence similarity to MiaB, which methylthiolates tRNA. We show that RimO and MiaB represent two of four subgroups of a larger, ancient family of likely methylthiotransferases, the other two of which are typified by Bacillus subtilis YqeV and Methanococcus jannaschii Mj0867, and we predict that RimO is unique among these subgroups in its modification of protein as opposed to tRNA. Despite this, RimO has not significantly diverged from the other three subgroups at the sequence level even within the C-terminal TRAM domain, which in the methyltransferase RumA is known to bind the RNA substrate and which we presume to be responsible for substrate binding and recognition in all four subgroups of methylthiotransferases. To our knowledge, RimO and MiaB represent the most extreme known case of resemblance between enzymes modifying protein and nucleic acid. The initial results presented here constitute a bioinformatics-driven prediction with preliminary experimental validation that should serve as the starting point for several interesting lines of further inquiry.
Proc Natl Acad Sci U S A
2008;105(6):1826-1831
| PubMed ID:
18252828
Landgraf BJ, Arcinas AJ, Lee KH, Booker SJ
Identification of an Intermediate Methyl Carrier in the Radical S-Adenosylmethionine Methylthiotransferases RimO and MiaB
▸ Abstract
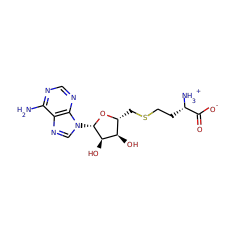
RimO and MiaB are radical S-adenosylmethionine (SAM) enzymes that catalyze the attachment of methylthio (-SCH3) groups to macromolecular substrates. RimO attaches a methylthio group at C3 of aspartate 89 of protein S12, a component of the 30S subunit of the bacterial ribosome. MiaB attaches a methylthio group at C2 of N(6)-(isopentenyl)adenosine, found at nucleotide 37 in several prokaryotic tRNAs. These two enzymes are prototypical members of a subclass of radical SAM enzymes called methylthiotransferases (MTTases). It had been assumed that the sequence of steps in MTTase reactions involves initial sulfur insertion into the organic substrate followed by capping of the inserted sulfur atom with a SAM-derived methyl group. In this work, however, we show that both RimO and MiaB from Thermotoga maritima catalyze methyl transfer from SAM to an acid/base labile acceptor on the protein in the absence of their respective macromolecular substrates. Consistent with the assignment of the acceptor as an iron-sulfur cluster, denaturation of the SAM-treated protein with acid results in production of methanethiol. When RimO or MiaB is first incubated with SAM in the absence of substrate and reductant and then incubated with excess S-adenosyl-l-[methyl-d3]methionine in the presence of substrate and reductant, production of the unlabeled product precedes production of the deuterated product, showing that the methylated species is chemically and kinetically competent to be an intermediate.
J Am Chem Soc
2013;135(41):15404-15416
| PubMed ID:
23991893
Landgraf BJ, Booker SJ
Stereochemical Course of the Reaction Catalyzed by RimO, a Radical SAM Methylthiotransferase
▸ Abstract
RimO is a member of the growing radical S-adenosylmethionine (SAM) superfamily of enzymes, which use a reduced [4Fe-4S] cluster to effect reductive cleavage of the 5' C-S bond of SAM to form a 5'-deoxyadenosyl 5'-radical (5'-dA(•)) intermediate. RimO uses this potent oxidant to catalyze the attachment of a methylthio group (-SCH3) to C3 of aspartate 89 of protein S12, one of 21 proteins that compose the 30S subunit of the bacterial ribosome. However, the exact mechanism by which this transformation takes place has remained elusive. Herein, we describe the stereochemical course of the RimO reaction. Using peptide mimics of the S12 protein bearing deuterium at the 3 pro-R or 3 pro-S positions of the target aspartyl residue, we show that RimO from Bacteroides thetaiotaomicron (Bt) catalyzes abstraction of the pro-S hydrogen atom, as evidenced by the transfer of deuterium into 5'-deoxyadenosine (5'-dAH). The observed kinetic isotope effect on H atom versus D atom abstraction is ∼1.9, suggesting that this step is at least partially rate determining. We also demonstrate that Bt RimO can utilize the flavodoxin/flavodoxin oxidoreductase/NADPH reducing system from Escherichia coli as a source of requisite electrons. Use of this in vivo reducing system decreases, but does not eliminate, formation of 5'-dAH in excess of methylthiolated product.
J Am Chem Soc
2016;138(9):2889-2892
| PubMed ID:
26871608
Molle T, Moreau Y, Clemancey M, Forouhar F, Ravanat JL, Duraffourg N, Fourmond V, Latour JM, Gambarelli S, Mulliez E, Atta M
Redox Behavior of the S-Adenosylmethionine (SAM)-Binding Fe-S Cluster in Methylthiotransferase RimO, toward Understanding Dual SAM Activity
▸ Abstract
RimO, a radical-S-adenosylmethionine (SAM) enzyme, catalyzes the specific C3 methylthiolation of the D89 residue in the ribosomal S12 protein. Two intact iron-sulfur clusters and two SAM cofactors both are required for catalysis. By using electron paramagnetic resonance, Mössbauer spectroscopies, and site-directed mutagenesis, we show how two SAM molecules sequentially bind to the unique iron site of the radical-SAM cluster for two distinct chemical reactions in RimO. Our data establish that the two SAM molecules bind the radical-SAM cluster to the unique iron site, and spectroscopic evidence obtained under strongly reducing conditions supports a mechanism in which the first molecule of SAM causes the reoxidation of the reduced radical-SAM cluster, impeding reductive cleavage of SAM to occur and allowing SAM to methylate a HS(-) ligand bound to the additional cluster. Furthermore, by using density functional theory-based methods, we provide a description of the reaction mechanism that predicts the attack of the carbon radical substrate on the methylthio group attached to the additional [4Fe-4S] cluster.
Biochemistry
2016;None(None):None-None
| PubMed ID:
27677419